|
DNA Technology DNA is noting new, in fact it has been the subject of research since the first published paper back in 1871. But the technology to use DNA for identification was not available until over 100 years later in 1986 and then made commercially available in 1987. Here we are almost 30 years later and technology has come a long way since 1987. We have evolved from rotary phones to Internet capable cell phones, computers in every home and cars that can drive themselves. Of course DNA technology has also advanced making genetic profiling more attainable and less expensive.
There is still a long way to go and advances in DNA technology emerge daily, but at DOGdex we think we have reached a point where the technology can be used to save dogs lives and get them returned home when lost. So how does this work? How can DNA help someone identify their dog when lost? The process is very basic. A cotton swab is used to take a saliva sample from each dog in a municipality, then when dogs are lost, a sample is taken from the lost dog and the sample is matched against all the existing samples to find the actual owners. As long as you have an ID for your dog before they are lost then the dog can be returned to you. Of course the actual DNA part is a little more complex. How DOGdex ID DNA Works in a Nutshell The process of actually matching the DNA is extensive and requires many steps. Obviously, we can't hold up the two cotton swabs to a light bulb to see if they match. A single strand of dog DNA is made up of billions of nucleotides and even with the variance in breeds, sizes and appearances 99.9% of the DNA will be the same, however, there are significant variations in particular sequences that have been isolated as known genetic markers.
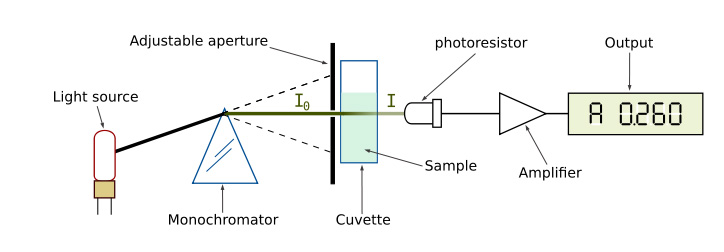
We use 23 of those genetic markers to create an identity for each dog, which much greater than the 14 the genetic markers used by the FBI in human cases for DNA matching. We need to isolate just those genetic markers which are short sequences, typically only around 20 nucleotides, and match the size fragments of those strands with all the sizes of all existing profiles. First the DNA is collected using a simple cotton swab to gather some saliva from the dogs mouth. That sample is then put through an extraction or lysis and purification process which separates out just the DNA from the sample and removes impurities that would inhibit the analysis. The process involves several steps of heating, cooling and filtering using a centrifuge. Once the DNA is isolated it is quantified to determine if an adequate amount of DNA was collected. The sample is diluted in a cuvette and placed in a spectrophotometer which uses light at a specified wavelength to measure the density of the concentration in the sample as shown below.
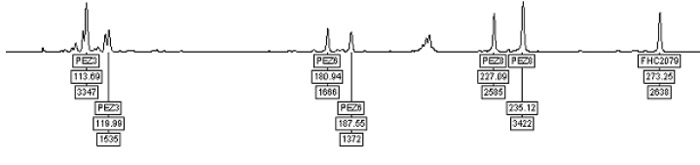
Then forward and reverse oligos nucleotide primers are added which will allow us to duplicate only the genetic markers that are needed for identification rather than the entire strand of those billions of nucleotides. The amplification is done using a thermocycler in a process known as polymerase chain reaction or PCR. The thermocycler slowly heats and cools the DNA for several hours which causes it to replicate until there are billions of duplicate strands of the isolated genetic markers. Of course, even with billions of strands they are so small they are contained in less than a single drop of water. The amplified DNA, that single drop, is then analyzed using capilliary electrophoresis which uses a electrical charge to draw the DNA samples through hair sized capillaries filled with a liquid polymer. The size of each marker is determined by the time that it takes to travel through the capillary. Below is a sample of the data provided by the capilliary electrophoresis.
For each maker there are 2 Allleles represented by peaks on the graph, one is inherited from each parent. This genetic map is our digital fingerprint that we can compare with other digital fingerprints. Comparison is done by constructing datasets of all 23 genetic markers including the 14 which appear on all of the DOGdex IDs. In the sample above the marker PEZ03 shows relative values of 113 and 119 and marker PEZ05 with relative values of 180 and 187 which all become solid data points for use in identification. Once we have converted that size information into digital data we can compare that data with existing profiles to find a match. We have developed software not unlike that used by the FBI which interprets those data points and finds likely matches. Then the actual data can be reviewed by technicians to validate the match. The process works very well and has been used for many years but the science is complex and the process is difficult and requires very sensitive specialized equipment. While anyone can collect samples from their dog, only extremely qualified people can run the analysis due to the sensitivity of the derived data and measuring instruments. This is not a simple or quick process as seen on TV, it is quite complex and the result of years of genetic research and development by commercial companies and Universities across the globe. The science is solid and being used by dog registries and research teams. We have taken input from hundreds of studies and streamlined the process to make it affordable and cost effective bringing the end product to the consumer for under $35. When we have this type of technology at our fingertips at such an affordable price, there is no excuse for euthanizing over a million dogs a year when the owners want them back.
|